
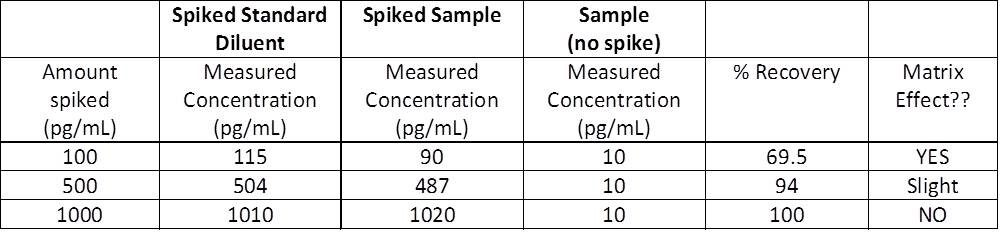
This should be an essential component of your experiment. We recommend including an endogenous positive control if you are testing a recombinant protein sample. This is known as a spike control and tells you that a target protein is recoverable after being spiked into a matrix. The two can then be compared to ensure there is no effect on the standard curve from other proteins in the serum. But we recommend also including a standard diluted in serum from the species you are testing. When testing serum samples in ELISA, include a standard in normal diluent buffer as usual. Standard in sample matrix (spike control) control The R 2 value of the trend line should be >0.99. A poor standard curve means the antibody didn’t bind properly or doesn’t capture the protein standard. For example, below is a typical standard curve from our Murine IL6 ELISA kit with concentration ranging from 15.6 to 500 µg/mL. This is a sample that contains a known concentration of the target protein from which the standard curve can be obtained. Each plate you use should contain a negative control sample in order to validate the results. This is to check for non-specific binding and false positive results. This is a sample that you know does not express the protein you are detecting. If you still have difficulty finding a suitable control, we recommend doing a quick literature search on PubMed to see which tissues and cells express the protein of interest.This will usually provide you with relative levels of expression in various tissues. Check the GeneCards entry for the protein.These can also be considered suitable positive controls. These databases will often have a list of tissues that the protein is expressed in. Try looking at the Swiss-Prot or Omnigene database links on the datasheet.Check to see if there are any Abreviews® for the antibody. Any tissues, cells, or lysates that have been used successfully can be considered a suitable positive control.If no control is suggested, we recommend the following: We recommend checking the antibody datasheet, which will often provide a suggested positive control.
It will verify that any negative results are valid. A positive result from the positive control, even if the samples are negative, will indicate the procedure is optimized and working. Use either an endogenous soluble sample known to contain the protein you are detecting or a purified protein or peptide known to contain the immunogen sequence for the antibody you are using.


 0 kommentar(er)
0 kommentar(er)
